Setting the Scene: The Tools and the Data
The Hardware
When we at the Danish State and University Library (SB) started our work on the SCAPE project, we acquired four machines to support this work. It was on those four machines that the FITS experiment was performed and it is on the same four machines that the present experiment was performed. The big difference being the software that handles the processes and how the data is accessed.
The four machines are all Blade servers each with two six core Intel Xeon 2.93GHz CPUs, 96GB RAM, and 2Gbit network interfaces (details at http://wiki.opf-labs.org/display/SP/SB+Test+Platform). Much to the contrary of a traditional node for a Hadoop cluster, these machines do not have local data storage. At SB we very much rely on NAS for data storage, more specifically our storage is based on the Isilon scale out NAS solution from EMC. The Isilon system is a cluster designed for storage and at SB it is at the present storing several PB of digital preservation data.
As NAS is a prerequisite for data storage at SB, we have been doing a lot of experiments on how best to integrate Hadoop with that kind of infrastructure. We have tested two Hadoop distributions, Cloudera and Pivotal HD, in different hardware and software configurations and the setup used for this experiment is the best so far while in no way being good enough. For the experiment we used the Cloudera distribution version 4.5.0, which builds upon Hadoop 2.0.0.
The Data
During the last months we have been moving our copy of the Danish Web Archive onto the above-mentioned Isilon cluster and providing online read-only access through NFS. This makes it possible for relevant jobs to access more than half a PB of web documents, roughly more than 18 billion documents harvested during the last decade. That is a lot of data!
These documents are stored as ARC or WARC container files in a shallow directory tree with the ARC and WARC files in the nethermost directory. A few months ago we shifted from storing data in the ARC format to storing the data in the newer WARC format as this format is superior to the older ARC format. This will become relevant later in this post.
To select the data for this large scale experiment, a few simple UNIX find commands were issued giving a file with the file paths of 147,776 ARC files amounting to almost 15TB, roughly around 450 million documents.
The Software
For this large-scale characterisation experiment I substituted FITS with the Nanite project. This project is lead by Andy Jackson from the UK Web Archive and enables DROID, Apache Tika, and libmagic to be effectively used on the Hadoop platform. For this experiment I will not use the libmagic component. Will Palmer of the British Library (BL) has written a blog post on this project (http://www.openpreservation.org/blogs/2014-03-21-tika-ride-characterising-web-content-nanite) and how to integrate the Apache Tika parser into Nanite. Will Palmer has also been very helpful with uncovering and rectifying the problems uncovered during the present experiment.
Before unleashing Nanite on the big data set, I did a lot of preliminary tests and tweaks in the source code of Nanite, more specifically the nanite-hadoop module that compiles to a JAR containing a self-contained Hadoop job. (In this blog post Nanite is a synonymy for nanite-hadoop)
One tweak was to write code for extracting the ARC metadata for each document and storing it along side the properties extracted by Tika.
Another tweak was to substitute the newlines in the extracted HTML title elements with spaces as those newlines breaks the data structure of one key/value pair per line in one of the output data files.
Both these two changes are now in the main Nanite code base.
After disabling a small hack that enables the code to run on the BL Hadoop cluster and disabling a preliminary check to ensure that all input files are compressed, which ours are not, the code should be ready for some real test runs.
Not quite so! The Nanite project depends upon another UK Web Archive project called warc-hadoop-recordreaders, which again depends on a version of the Heritrix project that has a bug when dealing with uncompressed ARC files. The Heritrix code simply cannot handle such uncompressed ARC files. When this bug was uncovered (with help from Will and Andy), it was easy fixing the problem by using a newer, but unreleased version of Heritrix (3.1.2-SNAPSHOT) and rebuilding the dependencies.
During some fun and interesting Skype conversations with Will, we decided to have three different output formats for Nanite. So, basically Nanite produces two kinds of outputs. It produces a traditional data set that are created by the mappers and aggregated in the reducers. This data set contains lines of MIME type, format version, and, for DROID, PUID values. The other kind of output is the extracted features of the documents. Each mapper task handles one ARC file at a time and creates a single data set with all the extracted features from all the documents contained in the ARC file. This data set is then stored by the mapper in three different containers.
- A ZIP file per ARC file that contains one file per document with key/value pairs, one per line.
- A ZIP file per ARC file containing the serialised metadata objects for each document.
- A sequence file per ARC files with the serialised metadata objects for each document.
When we know more about who will use this data and to what purpose, those three formats could be reduced to one or substituted by something entirely different, maybe even improving the performance of the process. The above three output formats are only available in Will’s fork of Nanite at https://github.com/willp-bl/nanite.
As a last experiment in this prologue I ran Nanite on 221GB of ARC files to test the performance. This test showed a processing speed at 4.5GB/minute. The test created circa 1GB of extracted metadata. Extrapolating these values it seemed that the complete 15TB could be processed in less than three days and giving less than 100GB of new data. The latter would have no problem in fitting on the available HDFS space and three days processing time would be impressive, to say the least.
Running the Experiment
Only thing left was to execute
$ hadoop jar jars/nanite-1.1.5-74-SNAPSHOT-job-will.jar ~/working/pmd/netarkiv-147776 out-147776
and wait three days.
I just forgot that this was big data. I forgot that with sheer data size comes complexity. That with 147,776 ARC files, something had inevitable to go wrong.
Before initiating this first run I had decided that I wouldn’t change code or fiddle with the cluster configuration more that absolutely necessary. My primary focus was to get the job to run by jumping as many fences as I had to get the job to terminate without failure.
So when I after a few seconds got a heap space exception, I didn’t change heap space configuration. Instead I split the input file into two equal sized chunks with the intention of running two separate Hadoop jobs.
When I subsequent got an “Exceeded max jobconf size” I didn’t experiment with the mapred.user.jobconf.limit Hadoop parameter but split the original input file into chunks with only 30,000 ARC references each.
After that I started getting “ZIP file must have at least one entry” exceptions. First I removed 4832 ARC files that didn’t contain documents but instead contained metadata regarding the original web harvest. I didn’t know these files were present until I got the above error. That didn’t fix the problem. Next I discovered that I unexpectedly also had WARC files in my data set and the code presumable couldn’t handle those. The set of 147,776 files was now reduced to 79,831, split into three chunks. As a last guard against the job failing I did something bad. I surrounded the failing code with a try-catch instead of understanding the problem—actually I just postponed the real bug-hunt for another opportunity.
Late Saturday evening the first job was about to complete, but as I didn’t want to stay up too late just for starting a Hadoop job, I dared to try to run two Hadoop jobs at the same time. Wrong decision! The second job started spitting out a lot of ”Error in configuring object”. So I killed it and went to bed. The next morning the first job had, luckily, completed with success and I had the first set of results. Off course I then started the second job yet again on a idle Hadoop cluster, but, very worrying, got the same error as in the previous evening! It being Sunday, I closed the ssh connection and tried to forget all about this for the rest of the weekend.
Monday morning at work this second job started without hick-ups and I still don’t know what went wrong!
The second job ran for ten hours, completing 29,999 ARC files out of 30,000 and then it failed and cleaned everything out. Upon examination of the log files and the input data I discovered an ARC file of size zero! That is a problem, not only for my experiment, but certainly also for the web archive and this discovery has been flagged in the organisation.
I removed the reference to this zero size ARC file, ran the job again with success. In the third job I also discovered two more zero size ARC files whose references were removed and the job completed with success.
After 32 hours of processing time, all 79,829 ARC files was processed and a lot of interesting metadata created for further analysis in addition to new experience and knowledge gained.
Analysis
As mentioned above the job produces two kinds of metadata, but it is actually three kinds: MIME type identifications of the documents stored in tab separated text files, extracted metadata for each document stored in ZIP files, and data about the job run itself. I will analyse the last kind first.
To count the number of processed documents I ran this Bash script
for f in $(cat all-files.txt)
do
rn=$(basename $f .zip)
s=$(ls -l $f|awk '{print $5}')
c=$(unzip -l $f|tail -1|awk '{print $2}')
echo "$f, $c, $s"
done
that creates a list of ARC file name and document count pairs. This data gives the total amount of processed documents, which were 260,603,467, and the following distribution of number of documents per ARC file
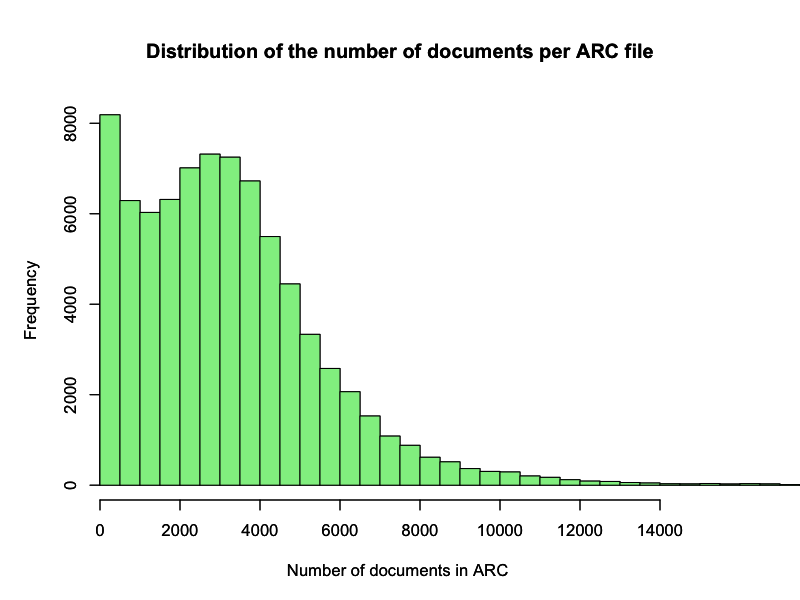
In the tail now shown we have 350 ARC files with more than 15,000 documents. The ARC file with the most documents counts 41,140 pieces. Again, an anomaly that would be interesting to dive into as well as many other questions this distribution rises.
The execution time for the three sub jobs were
| job id | amount of data read | number of ARC files read | processing time |
|---|---|---|---|
| 0059 | 2.764TB | 30,000 | 12hrs, 43mins, 53sec |
| 0069 | 2.763TB | 30,000 | 10hrs, 46mins, 21sec |
| 0073 | 1.820TB | 19,829 | 7hrs, 44mins, 38sec |
| 7.347TB | 79,829 | 31.24hrs |
Basically the cluster processed 3.92GB/minute, or 44 ARC files per minute, or 63,000 ARC files a day, or 2317 documents per second—which ever sounds most impressive.
To dig a bit deeper into this data set, I collected the run time of each of the 79,839 map tasks. This data can be explored in an HTML table on the job level in the Cloudera Manager, but I wrote a small Java job that converts this data into a CSV file: https://github.com/perdalum/extract-hadoop-map-data. The data was then read into R for analysis. For the interested reader, I created a Gist with the unordered R code that generated the analysis and figures at https://gist.github.com/perdalum/a9041ff3f245986a62f3
The processing time spans from -43s [sic] to 1462s. A very weird observation is that 6,490 tasks, i.e. 8%, were reported as having completed in negative time. As I don’t think the Hadoop cluster utilises temporal shifts, though that would explained the fast processing, this should be investigated further.
Only 83 map task took more than 500s. Ignoring that long thin tail, the distribution of the processing times looks as below
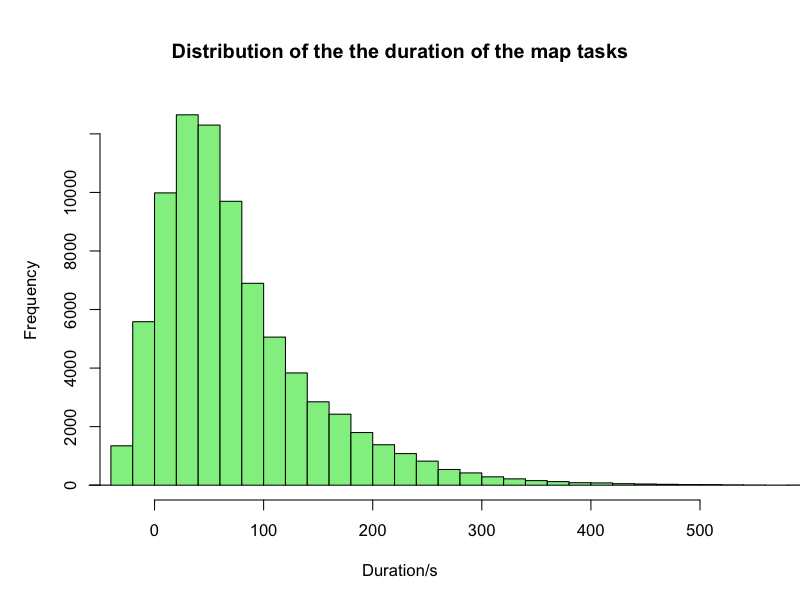
This looks like an expected distribution. The median of this data set is 57s but that includes 8% negative values so it’s unclear what valuable information that gives.
The MIME type identifications are aggregated in 10 files, one per reducer. These files are easily aggregated further into one big file. Eyeball examination of this data file reveals that it contains lots of error messages. The course of these errors should be investigated, and Will has been doing that, but I’ll jump the fence once more, this time using grep
grep -v ^Exception all-parts | grep -v ^IOException > all-parts-cleaned
Still, that was not enough because after trying to read the file into R, I discovered that I needed to remove all single and double quotes
sed 's/"//g' < all-parts-cleaned | sed "s/\'//g" > all-parts-really-cleaned
With that data cleaning completed, the following command finally reads the data into R
r<-read.table("data/all-parts-really-really-cleaned",sep="\t",header=FALSE, comment.char="",col.names=c("?","http","droid","tika","tikap","year","count"))
Observe the comment.char=”” argument. This is necessary as R assumes everything after a # is a comment and in three instances in the data an http server reported a MIME type value as a series of #s. Why? The same server? This just gives rise to even more interesting questions that can be asked about the uncovered anomalies in this data.
Apart from the above mentioned error messages, the data also contain another kind of error. The Tika parser has timed-out on a lot of the documents, 177 million of them, to be exact. That is a 67% failure rate for the Tika parser and I would consider that a serious error. Fortunately this error has already been dealt with by Will and pushed to the main Nanite project.
Had I chosen to include the harvest year into this data during the job, the detected MIME types could be correlated with harvest year. Instead I can compare the distribution of MIME types between the values reported by the server that served the documents, the DROID detection, and the Tika detection.
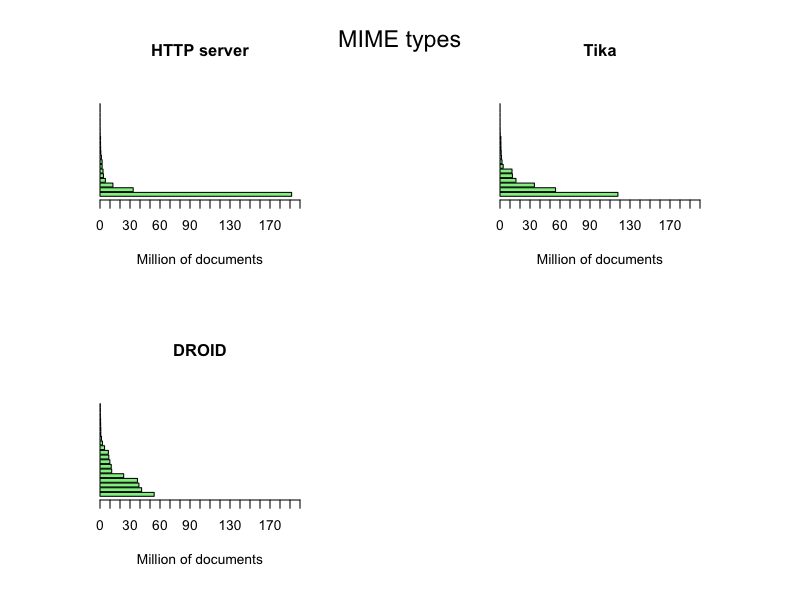
The web servers reported 1370 different MIME types; Tika detected 342; and DROID 319.
A plot of the complete set of these MIME types is presented for completeness

If we select the top 20 MIME types we get a more clear picture
Top 10 with MIME type names
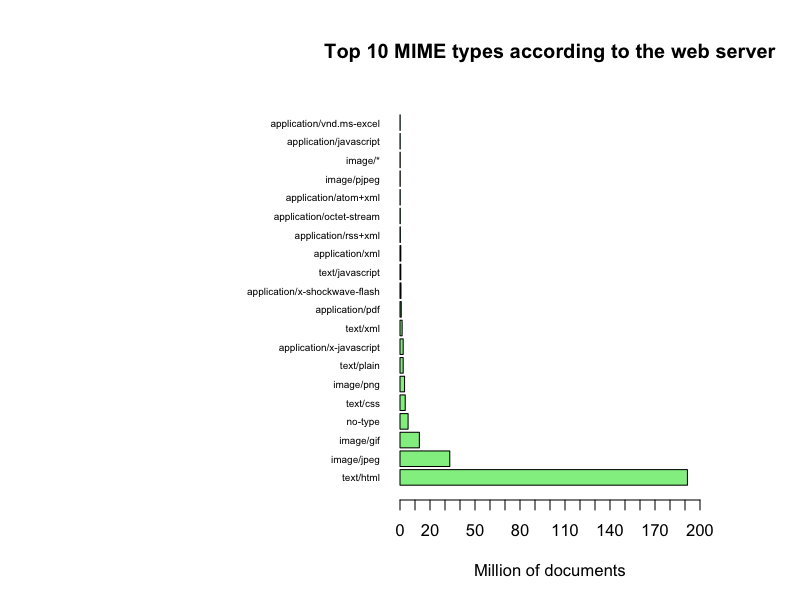
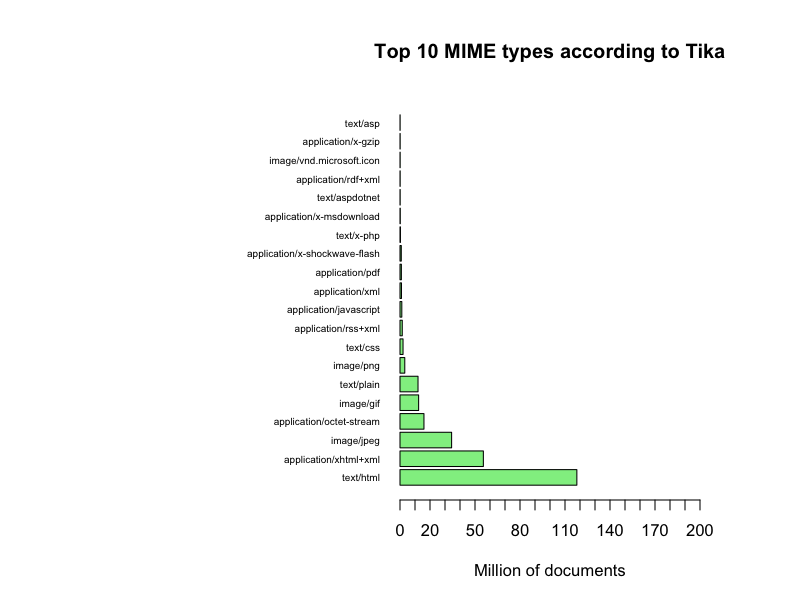
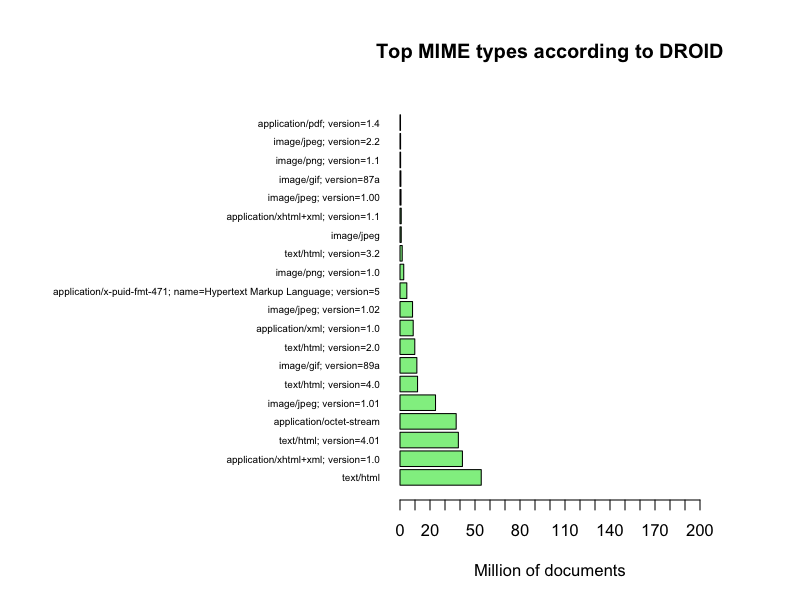
It seems that the web servers generally claim that they deliver a few documents with a lot of few different MIME types and a lot of documents with a few MIME types, primarily HTML pages, when in fact we receive quite a diversified set of document types as detected by Tika and DROID. Also, this data set can actually give a confidence factor for the trustworthiness the Danish web servers, even a time series plot of the evolution of such a confidence factor. Or…, Or… The questions just keep coming…
Manually examination of the MIME type data set also reveals fun stuff. E.g. how did a Microsoft Windows Shortcut end up on the Internet? Or what about doing a complete virus scan of all the documents. This could give data for research in the evolution of computer viruses?
There’s no end to the interesting questions when browsing this data. And that’s just looking at one feature, namely the MIME type.
The third kind of data created in the Hadoop job gives a basis for much more detailed observations. We’ve got EXIF data, including GPS if available, HTML title and keyword elements, authors, last modification time, etc. I selected at random a data file from a typical ARC file, i.e. one that contains circa the average of 3500 documents. This data file has 430 unique properties!
These extracted properties are collected in 79,829 ZIP files and before any analysis would be feasible, these files should be combined into one big sequence file and a few Pig Latin UDFs should be written. This would facilitate a kind of explorative analysis. How close to real-time read, eval, print, loop, such a investigation could be, remains to be seen, as this is, unfortunately, a task for another day.
Lessons learned
First off, thank you if you’ve read this far. I did put quite a bit of detail into this blog post—without any TL;DR warning.
The process of going from the idea of this experiment to finishing this blog post has been long but very rewarding.
I’ve uncovered bugs in and added features to Nanite in fun collaboration with Will Palmer and this is far from over. I will continue working with Will on Nanite as I see great value in this tool. As a start I would like to address all the issues uncovered as described in this blog post.
I’ve learned that CPU time actually is much cheaper than developer time. If the cluster should run for 10 hours and fail in the last few minutes, who cares? It’s better than me spending hours trying to dig up a small bug that might be hidden somewhere down in the deepest Java dependencies. Up till a certain break-even threshold, that is!
I’ve learnt a valuable lesson: Know thy data! When performing jobs that potentially could run for weeks, it is very important that you know the data you’re trying to crunch. Still, it’s equally as important to have your tools be indeed very robust. They should be able to survive anything that might be thrown at them, because, running on large data amounts like this, every kind of valid and non-valid data will be encountered. I.e. if one file in a million is a serious problem, you will encounter 17,000 serious problems processing the web archive. If the job runs for 3 months, that’s almost 10 serious problems an hour.
All this being said and done, we must not forget why we do this. It’s not for the sake of creating fast tools, nor reliable tools. It is to enable the curators to preserve our shared data as easy and trustworthy as possible. Also, especially relevant for web documents, it’s for the benefit of the researches in the humanities. To enable them to get answers to all the different questions they could possible imagine asking to such a huge corpora of documents from the last decade.
Even though this experiment answered some questions, I now stand with even more questions to be answered. Oh, and I need to run Nanite on 18 billion documents that are just waiting for me on a NFS mount point…



May 29, 2014 @ 10:35 am CEST
It's great to see this kind of detailed write-up on this subject, and I'm glad to see Nanite getting a work-out on your data. Here at the UK Web Archive, we've integrated it into our full text search stack, and are looking at ways of making this kind of information more usable for curators and researchers by exposing it as Solr search facets. Thanks to the work by yourself and Will, I know that SCAPE Nanite should be robust enough for us to depend on.