In the previous blogpost we focussed on the preservation of Virtual Machines. In the second part we focus on the preservation of a computational scientific workflow implemented in a common workflow engine as a software container.
Container technology (also called operating system virtualization) is promoted as a lightweight alternative to virtual machines, requiring less resources while maintaining portability. Driven by the demand for reproducible computational science, containers e.g. Docker and in the scientific communities esp. Singularity have been adopted quickly by researchers, as containers are able to encapsulate a software environment (e.g. a complex software tool-chain including application-specific settings) into a single portable entity.
While containers offer a set of features convenient for the preservation of (complex) software setups, popular container implementations have been criticized for their poor backward compatibility. To make scientific methods accessible, usable and citable in the long-term, the longevity of containers as a new digital object class needs to be ensured.
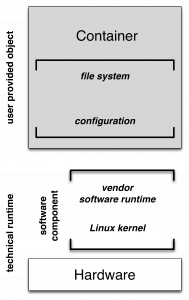
Due to the strict isolation (portability requirement), containers need to be self-contained, i.e. all software dependencies (libraries, applications etc.) have to be included within the container. Thus, the main component of a container is a self-contained file-system with installed and configured software components. The only remaining external or unresolved software dependency is the underlying operating system, i.e. the operating system’s application binary interface (ABI). The second container component is its runtime-configuration defining, for instance, file-system mappings, e.g. shared folders between container and its host system, and the definition of an entry-point within the container (i.e. a script or program).
A New Class of Digital Objects with Preservation Risks
Hence, a container’s technical runtime is composed of two components: a hardware component (the computer) and a software component. In general, the software component represent typically a basic Linux installation with an installed and configured (vendor specific) container runtime. Preserving containers and their technical runtime can be decoupled by
- creating and preserving generic container runtime setups
- and replacing outdated hardware with virtual or emulated hardware.
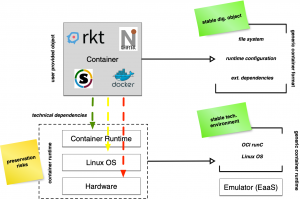
Even though containers build on the same technical foundations, currently popular container implementations such as Docker, Singularity or Shifter use different container representations and configuration formats. These representations typically require a vendor specific technical runtime. Ideally, a small number of software runtimes should be sufficient to run a large number of containers, ie to reduce future management and maintenance effort. Hence, a common container representation and especially configurations reduces the preservation risk to the availability of a common software runtime. All container formats and variations are based on the same technical foundations, vendor specific additions and features typically improve usability and simplify container development. As we assume, that containers subject to publication and archival are immutable, we decided to generalize containers by dissecting vendor specific components into a common, (partly) standardized structure, such that for an archive’s perspective all container “objects” share the same technical properties, ie. technical dependencies. Furthermore, we can assume that the operating system interface rarely changes in incompatible ways, such that only a few variations of such runtimes are necessary.
Example
This example has been prepared and described by Felix Bartusch (University Tübingen).
Scientific (software-based) workflows usually yield results that can be published as a paper in scientific journals. The authors of the paper have to describe their methodology used to compute their results in a way that other researchers are able to reproduce their findings. The description of methods and software in the paper is however not always sufficient to reimplement the whole workflow. In a case study researchers tried to reimplement a workflow used in a ‘typical computational biology paper’, which took them several months [1]. Their main problem was that not all important details were explicitly described in the paper and specific software versions were not available three years after the original publication. A potential solution for the reproducibility problems is the preservation of the workflow and its computational environment in a closed and well-formed unit like a container.
To illustrate how CiTAR works we use a tutorial workflow provided by the developers of OpenMS for the workflow engine KNIME (Konstanz Information Miner). OpenMS is a open-source software used to process mass spectrometry data. With KNIME one can create workflows by adding and connecting so-called nodes. Nodes provide configurable functions to process data in the workflow. The user can install additional nodes in order to add functionality to KNIME. The OpenMS developers created KNIME nodes that are extensively used in the example presented here.
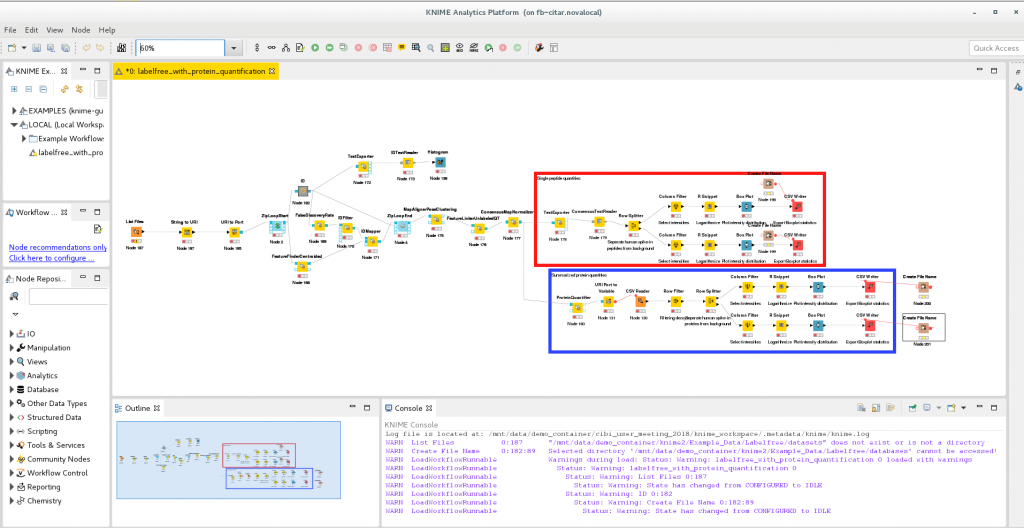
The software container submitted to the CiTAR service is created by using a experimental export functionality of the KNIME workflow engine that exports workflows together with their dependencies to software containers.
Submitting the Virtual Research Environment to CiTAR
We created such a container with the KNIME exporter and submitted it to CiTAR.
As there exist several containerization methods with different container file formats, CiTAR extracts the file system in the container and writes it to a disc image. The extracted file system on the disc image is now independent from a special container runtime. Thus CiTAR does not have to preserve many versions of different container runtimes, but uses the standardized runtime to execute a user defined process in the preserved environment.
Reproduce Results
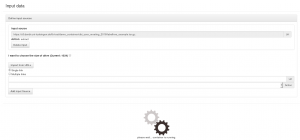
The preserved workflow can be executed on the container’s landing page.
The workflow performs a label-free protein quantification for mass spectrometry data of peptides. In a mass spectrometry analysis the peptides are ionized and accelerated, therefore travelling to the detector with a certain velocity. The time a peptide needs to reach the detector depends on the peptide’s mass and its charge. The raw data produced by such a mass spectrometer consists of signals at a specific Mass-to-Charge (m/z) ratio. The signal strength depends on the number of peptides in the sample. The research question is now: “How much of which protein was in the sample?”. Because the peptides were not labelled one needs to perform a ‘label-free’ protein quantification.
The workflow uses OMSSA, a mass spectrometry identification engine relying on a sequence database. The sequence database used in this example is also contained in the example dataset. It is a common step to not only search against a regular protein database, but to also search against a decoy database in order to perform false discovery rate (FDR) estimation and sort out unreliable results. The identified peptides are then quantified. They are currently just ion signals in the mass spectrometry data. A signal is not one specific value, but rather a scattered around a small portion of the m/z axes with a peak. The FeatureFinderCentroided node finds and quantifies these peptide ion signals contained in the data.
The example data provided in the OpenMS tutorial consists of three files, each with the same level of background peptide noise but different levels of spiked-in human peptides. To show this, the workflow returns some numbers describing the distribution of background peptides and actual human peptides in each sample.
References
[1] Garijo, D., Kinnings, S., Xie, L., Xie, L., Zhang, Y., Bourne, P. E., & Gil, Y. (2013). Quantifying reproducibility in computational biology: The case of the tuberculosis drugome. PLoS ONE, 8(11), 1–11. https://doi.org/10.1371/journal.pone.0080278
The CiTAR Team
Rafael Gieschke, Susanne Mocken, Klaus Rechert, Oleg Stobbe, Oleg Zharkov (University Freiburg), Felix Bartusch (University Tübingen), Kyryll Udod (University Ulm)

